Visual ALP





Purpose
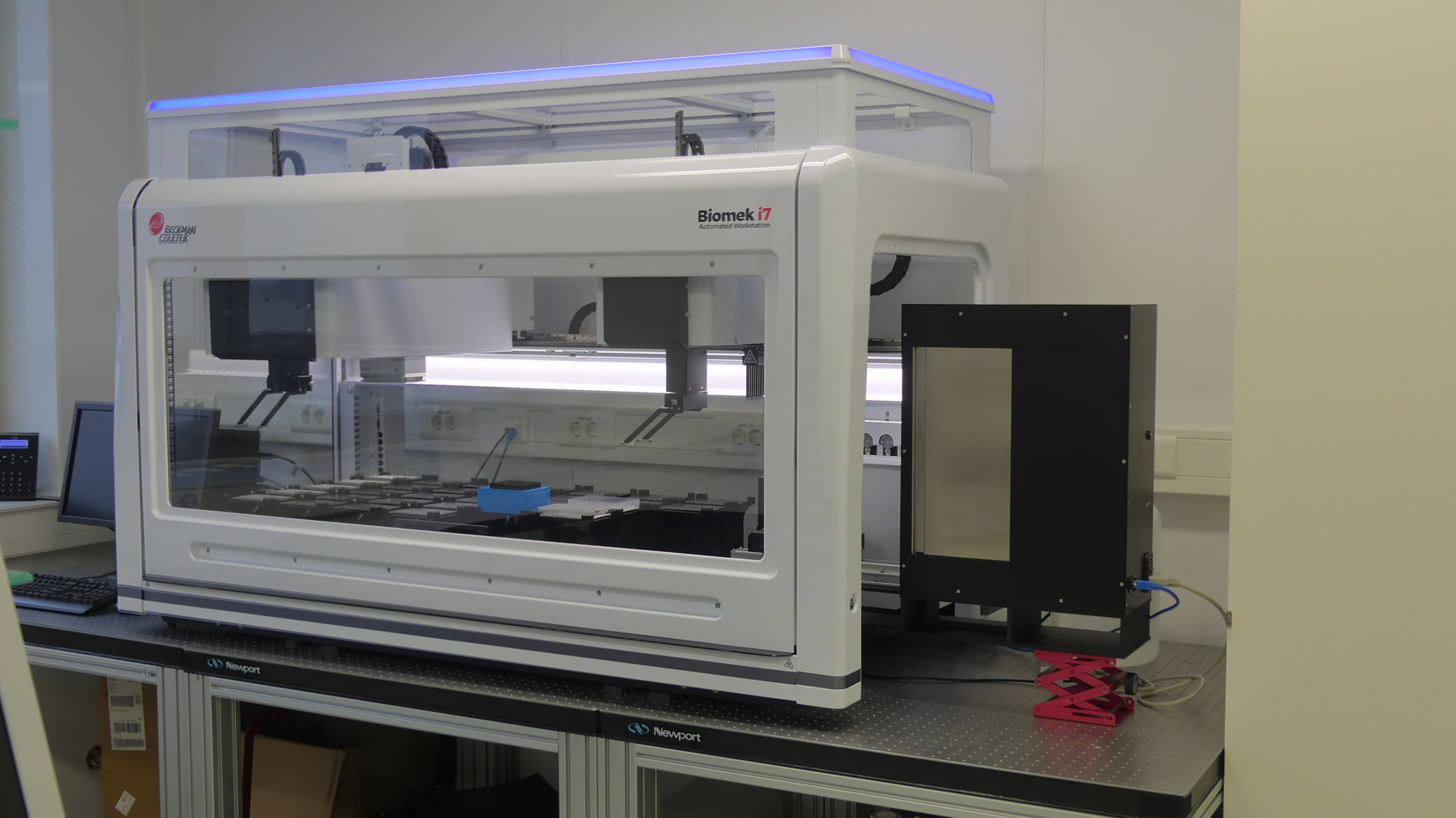
Cell line development plays a central role of any biologically-produced therapeutic to create stable producing cell lines for research and production. The most labour intensive step of the process is selecting the optimal clones from a pool of randomly generated cell lines. Single clones have to be picked for upstream and downstream processes.
A further application, the blue–white screening, allows the rapid and convenient detection of recombinant bacteria colonies often used in synthetic biology (synbio) workflows , white biotechnology or fundamental research. Cells transformed with vectors containing recombinant bacteria will produce white colonies; cells transformed with non-recombinant plasmids grow into blue colonies.
The Visual ALP was designed for a wide range of visual inspections including detection of colonies, blue-white screens, indicator colour detection etc. in Petri dishes and microtiter plate SBS format labware.
Details
The labware with colonies is illuminated from bottom with adjustable light (intensity, red / green / blue components). Illumination from top is optional. The images taken by a CCD camera are processed by the colony detection software allowing selection of colonies according to various parameters.
The Colony Editor program is used take or load images, to adjust detection and to select parameters to give best colony detection results. These parameters include camera and light settings. Detection images and detection results can be stored for post-process manual inspection.
Parameters and methods:
selection of colonies:
- Minimal and maximal size
- Minimal and maximal roundness (shape)
- Colour – minimal and maximal value for red, blue and green
minimal and maximal distance between colonies
methods for colony detection:
- Edge detection + shape separation
- Histogramm normalization + Threshold for foreground / background separation
- Dynamic edge detection + shape separation
- Detection area inside the labware wells
The coordinates and number of the selected colonies per labware well are provided to liquid handler software for further processing like colony picking, well selection etc. The duration of picking process depends on the number of detected colonies and on the technique of picking.
The colony detection for one Petri dish, MTP ot Omni trays is below 10 seconds including stage motion. Biomek speed performance for picking is dependent from internal settings and method structure. In a test method the Biomek takes about 40 seconds for 8 picks incl. dispensing into destination labware and tip loading / discard.
The Colony Detection Module was developed for the Biomek series from Beckman Coulter. The Visual ALP is completely integrated in the Biomek 5 Software and SAMI 4.x and 5.x.
It can be adapted to other liquid handler.